Why?
Membrane proteins mediate communication between the outside and inside of cells, and are common drug targets. Detailed understanding of the structure and function of these proteins enrich our basic understanding of how biology works at a molecular level, and can in the long run inform drug development. We have focused on the pentameric ligand-gated ion channels, which are a family of membrane proteins that respond to external chemical signals through subtle conformational changes that ultimately lead to the opening of a channel that allows ions to pass the membrane. While atomic resolution structures of these proteins have been highly informative in understanding this process, they do not paint the full picture as only a limited set of conformations are available. More information about e.g. structural dynamics at room temperature, and intermediate states, is needed for a more complete understanding of the gating landscape. It is possible to obtain low resolution structural information from proteins in solutions through small-angle scattering, so we set out to apply this technique to elucidate the solution structure of a ligand-gated ion channel, specifically GLIC, which is a bacterial protein often used as model system for these channels.
How?
We collected SANS data from GLIC at the small-angle diffractometer D22 at Institut Laue-Langevin, utilizing detergent deuterated to match the scattering length of pure heavy water (Midtgaard et al., 2018), allowing us to collect scattering data representing only the protein while maintaining maximal contrast between the protein and the buffer. To avoid the presence of protein aggregates in the measured samples we utilized the size-exclusion coupled setup at D22. Scattering was measured from GLIC under two buffer conditions; resting conditions (pH 7.5) where the channels are closed, and activating conditions (pH 3) where it is expected that a population of open channels will be present. Both conditions yielded overall similar scattering profiles, which when fitting a linear combination of the closed and open crystal structures revealed that the data from resting conditions were best fit by the closed structure alone, while the data from activating conditions gave equally good fits up to 18% contribution from the open structure. This value is close to a recent estimation of the open probability of GLIC (Bergh et al., 2021). To further improve our representation of the average solution conformations of GLIC we performed molecular dynamics simulations, which were launched from intermediate conformations obtained from elastic network extrapolations between the closed and open crystal structures. This gave us sufficient conformational sampling to identify conformations with improved goodness of fit to the SANS data, Figure 1A and B. We could thus conclude that while the average solution structure of GLIC at resting conditions is similar to the closed crystal structure, at activating condition it is best described by an intermediate conformation. Figure 1C.
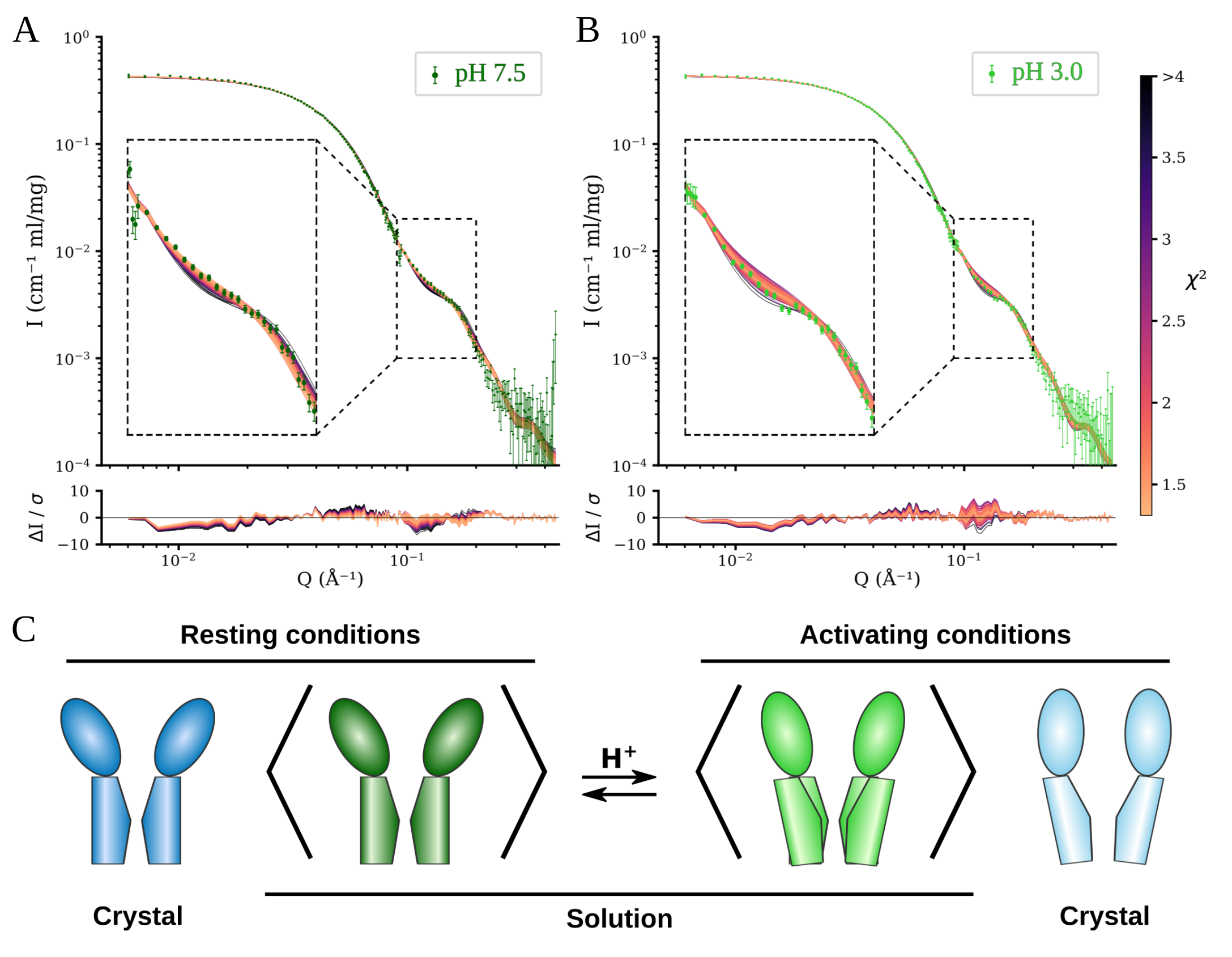
Figure 1. A) and B) Fits of models from molecular dynamics simulations to scattering data from resting conditions (A) and activating conditions (B). Models colored by goodness of fit. Lower panels show the error weighted residual between the models and the experimental data. C) Schematic comparison between the open and closed crystal structures of GLIC with the solution average conformation at resting and activating conditions. Adapted from Lycksell et al., 2021
References:
Bergh et al. Markov state models of proton-and pore-dependent activation in a pentameric ligand-gated ion channel. Elife 10 (2021): e68369.
Midtgaard et al. Invisible detergents for structure determination of membrane proteins by small‐angle neutron scattering. The FEBS journal 285, no. 2 (2018): 357-371.
What´s next?
With this work we have through a combination of SANS and molecular dynamic simulations been able to detect and describe small, condition dependent, changes in average solution structure. Having demonstrated this for GLIC, our next target is a related channel for which we aim to combine SANS, molecular dynamics simulations, and cryo-electron microscopy structure determination.
Who?
This work was lead by Marie Lycksell, PhD student in Erik Lindahl’s group, Stockholm University, and done together with Urška Rovšnik (Stockholm University), Cathrine Bergh (KTH Royal Institute of Technology), and Rebecca J. Howard (Stockholm University), Nicolai T. Johansen and Lise Arleth at the Niels Bohr Institute (University of Copenhagen, Denmark), and Anne Martel and Lionel Porcar at Institut Laue-Langevin (Grenoble, France). The project was supported by beamtime from Institut Laue-Langevin, computational resources from the Swedish National Infrastructure for Computing, and grants from SwedNess, the Knut and Alice Wallenberg Foundation, Swedish Research Council (2017-04641, 2018-06479, and 2019-02433), Swedish e-Science Research Centre, BioExcel Center of Excellence (EU 823830), and the Lundbeck Foundation through the Brainstruc grant.
Cite: Lycksell et al. Probing solution structure of the pentameric ligand-gated ion channel GLIC by small-angle neutron scattering. Proceedings of the National Academy of Sciences 118, no. 37 (2021): e2108006118
For the original articles, please visit:
https://doi.org/10.1073/pnas.2108006118
Contact:
Marie Lycksell
PhD student
Biochemistry and Biophysics, Stockholm University
Email: marie.lycksell@dbb.su.se
