Why?
The COVID 19 pandemic had worldwide impact in 2020 and continues to do so to this day. Understanding how the SARS-CoV-2 virus interacts with the body and why it has greater impact on some areas of the population than others is of great importance. Early on, studies found the need for the presence of cholesterol for the entry of the SARS-CoV-2 Spike (S) protein into cell membranes. Importantly, lower plasma total cholesterol, HDL and LDL levels, correlated to increased risk for severe disease. Moreover, both cholesterol and HDL were found to bind to the S protein. One of the roles of HDL is to remove excess cholesterol from cell membranes for elimination in the liver, and these results together suggest that HDL functionality might be affected by the interaction with the S protein. In this study we investigated the role of the SARS CoV-2 Spike protein on model membranes and its interaction with HDL by neutron reflection thereby offering insight into the role these components play in the progression of this disease.
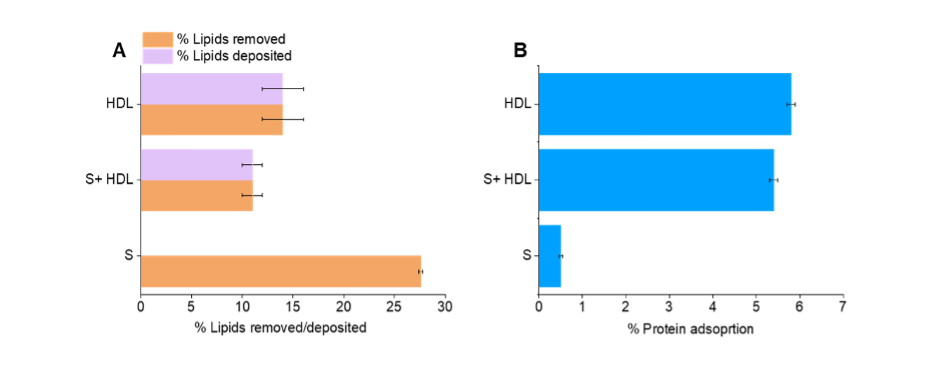
Figure 1. Quantities of A, lipids removed and deposited and B, protein adsorption to the membrane surface during the incubation with the S protein and/or HDL.
How?
Recently, we showed that neutron reflection could determine the extent of cargo deposition and lipid removal by HDL on model membranes. For this lipid deuteration was key, with the D-lab at the ILL providing both matchout deuterated natural lipids and matchout deuterated cholesterol. We then applied the same methodology to investigate the ability of Spike protein modify lipid exchange by HDL detailed here. Experiments were carried out on the D17 instrument - a reflectometer perfect for this type of experiment. Model membranes were prepared as supported lipid bilayers (SLBs) made of both saturated lipids and cholesterol. The membranes were characterised in three isotopic contrasts to compare membrane structure before and after incubation. This enables us to determine the processes which occurred during this time: lipid deposition, lipid removal, S protein and/or HDL adsorption (Figure 1).
Three independent membranes were prepared in this way. The first was incubated with S protein, the second was incubated with HDL and the third with a mixture of HDL and S protein. Neutron Reflection data were collected at different time points during these incubations (Figure 2). Progressive decrease in reflectivity in H2O based contrast over time indicates a reduction in the quantity of deuterated material adsorbed at the surface.
The S protein removed a portion of the membrane, however the quantity of lipids removed by HDL in the presence of S protein was only slightly lower than for HDL alone, suggesting that the combined effect of S on lipid removal was not the result of simply an additive effect by S and HDL. This suggests that indeed the S protein interacts with HDL, which probably leads to the S protein being saturated of HDL lipids and might lead to HDL composition remodelling.
Figure 2. Schematics of Spike protein and/or HDL incubation with model membranes, and corresponding graphs of lipid removal (red) and protein deposition (blue) over time. A, Spike protein alone, B, Spike protein with HDL and C, HDL alone incubations.
What´s next?
Neutron reflectometry has proven to be an excellent tool to study these systems providing clear insight into how the S protein interacts with model membranes and how it affects HDL function by reducing the quantity of lipids removed and deposited from the membrane, likely by removing lipids from the HDL itself and remodelling its composition and structure. Further studies to map HDL functionality behaviour and HDL remodelling by S protein could potentially lead to a better understanding of COVID-19 disease.
Who?
This work resulted from an international and cross disciplinary collaboration between the group of Prof. Marité Cárdenas at Malmö University and both life science and large scale sciences groups at Institute Laue Langevin, the Partnership of Structural Biology in France and the Austrian Center of Industrial Biotechnology.
Cite: J Colloid Interface Sci. 2021 Nov 15;602:732-739
For the original articles, please visit:
https://pubmed.ncbi.nlm.nih.gov/34157514/
Contact:
Prof. Marité Cárdenas
Department for Biomedical Science, Malmö University, Sweden
Email: marite.cardenas@mau.se
