Coupling lipid nanoparticle structure and automated single particle composition analysis to design phospholipase responsive nanocarriers
Why?
Lipid nanoparticles (LNPs) are currently under intense scrutiny for vaccine delivery and RNA therapies. Key to their successful optimisation and translation is a nanoscale, single particle characterisation of the relationship between LNP structure, composition, and performance. Maximising performance and understanding intracellular trafficking are key components of LNP rational design. Phospholipases, specific lipid modifying enzymes, are mediators of intracellular trafficking and therefore it is important to understand their ability to alter the LNP lipid composition. There are limited methods to characterise individual LNP composition, sample homogeneity and morphology. We present data relating LNP composition and structure to demonstrate phospholipase-mediated composition alterations in stable LNPs via both bulk and single LNP analysis. Through direct comparison of enzyme activity on vesicle and LNP structures, this work demonstrates the importance of multiscale characterisation and LNP lipid membrane structure for stimuli-responsive design of lipid nanocarriers.
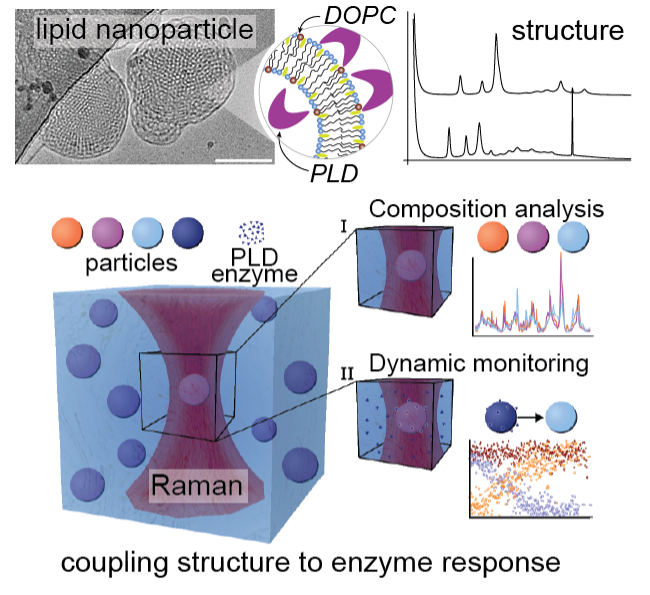
How?
Our multidisciplinary approach to composition and structure analysis of LNPs uses small angle scattering (SAXS and SANS) coupled with our Single Particle Automated Raman Trapping Analysis (SPARTAâ) technology, to relate lipid membrane structure and composition to specific phospholipase D-mediated alterations of the LNP lipid composition. Using neutrons, we were able to quantify changes in the LNP internal structure as a function of lipid composition, temperature and salt concentration. Our data showed that tuning the electrostatic interactions within our LNPs via charged lipids and NaCl enabled us to control the dimensions of the internal LNP structure. Interestingly LNPs generated with lipid compositions matching those produced by LNP – phospholipase D interactions were not structurally identical highlighting that formulation procedure is another key parameter that impacts LNP structure. Our structural data was combined with SPARTA to analyse the lipid compositions of hundreds of single LNPs. Our results showed that phospholipase D can access lipids from the entire LNP enabling full conversion of the lipid substrate to product and generating stable LNPs containing anionic lipid. The phospholipase D activity differs by an order of magnitude between LNP and vesicle structures, implying that lipid membrane curvature regulates phospholipase D activity and that LNP structure could be tailored to mediate interactions with phospholipases. This represents an opportunity to design enzyme-responsive LNP solutions for stimuli-responsive delivery and diseases where PLD is dysregulated.
What´s next?
In this work we measured an order of magnitude difference in phospholipase D activity between vesicles and structured LNPs. In future we are interested in understanding this interaction further to investigate if lipid membrane curvature can be used to control phospholipase D activity. Longer term this could enable the design of enzyme responsive pharmaceuticals.
Who?
This work stemmed from an international collaboration between researchers based at Karolinska Institutet (Sweden), Imperial College London (United Kingdom) and the ISIS Neutron and Muon Source (United Kingdom). Funding was provided via H2020 (Marie Skłodowska-Curie Fellowship “SmartCubes” (703666)), the Whitaker International Program, Institute of International Education, USA, FP7 (Marie Curie Intra-European Fellowship “SMase LIPOSOME” (626766)), the Swiss National Science Foundation (P300PA_171540), the NanoMed Marie Skłodowska-Curie ITN from the H2020 programme (676137), the Ermenegildo Zegna Founder’s Scholarship program, the Rosetrees Trust, the Engineering and Physical Sciences Research Council (EPSRC) (EP/R00529X/1, EP/K031953/1, EP/K020641/1), the ERC Seventh Framework Programme Consolidator grant “Naturale CG” (616417) and the Swedish Foundation of Strategic Research through the Industrial Research Centre “FoRmulaEx” (IRC15-0065). We acknowledge Diamond Light Source (beamline I22, SM23642, SM18658) for the award of synchrotron beamtime. Experiments at the ISIS Neutron and Muon Source were supported by a beamtime allocation from the Science and Technology Facilities Council (RB1810329). This work benefited from the use of the SasView application, originally developed under NSF award DMR-0520547. SasView contains code developed with funding from the European Union’s Horizon 2020 research and innovation programme under the SINE2020 project, grant agreement 654000. The Cryo-EM Swedish National Facility is funded by the Knut and Alice Wallenberg, Family Erling Persson and Kempe Foundations, SciLifeLab, Stockholm University and Umeå University.
Contact:
Prof. Molly Stevens
Department of Materials, Department of Bioengineering and Institute of Biomedical Engineering, Imperial College London, London SW7 2AZ, U.K. and Department of Medical Biochemistry and Biophysics, Karolinska Institutet, SE-171 77 Stockholm, Sweden.
Email: m.stevens@imperial.ac.uk
Dr. Hanna Barriga
Department of Medical Biochemistry and Biophysics, Karolinska Institutet, SE-171 77 Stockholm, Sweden
Email: hanna.barriga@ki.se
Cite: Adv Mater 2022, 34(26), e2200839